
Build a powerful question answering bot with Amazon SageMaker, Amazon OpenSearch Service, Streamlit, and LangChain
https://your-opensearch-domain-endpoint/llm_apps_workshop_embeddings/_count The browser window should show an output similar to the following… Run the following commands on the terminal to clone the code repository for this post and install the Python packages needed by the appl…
One of the most common applications of generative AI and large language models (LLMs) in an enterprise environment is answering questions based on the enterprise’s knowledge corpus. Amazon Lex provides the framework for building AI based chatbots. Pre-trained foundation models (FMs) perform well at natural language understanding (NLU) tasks such summarization, text generation and question answering on a broad variety of topics but either struggle to provide accurate (without hallucinations) answers or completely fail at answering questions about content that they haven’t seen as part of their training data. Furthermore, FMs are trained with a point in time snapshot of data and have no inherent ability to access fresh data at inference time; without this ability they might provide responses that are potentially incorrect or inadequate.
A commonly used approach to address this problem is to use a technique called Retrieval Augmented Generation (RAG). In the RAG-based approach we convert the user question into vector embeddings using an LLM and then do a similarity search for these embeddings in a pre-populated vector database holding the embeddings for the enterprise knowledge corpus. A small number of similar documents (typically three) is added as context along with the user question to the “prompt” provided to another LLM and then that LLM generates an answer to the user question using information provided as context in the prompt. RAG models were introduced by Lewis et al. in 2020 as a model where parametric memory is a pre-trained seq2seq model and the non-parametric memory is a dense vector index of Wikipedia, accessed with a pre-trained neural retriever. To understand the overall structure of a RAG-based approach, refer to Question answering using Retrieval Augmented Generation with foundation models in Amazon SageMaker JumpStart.
In this post we provide a step-by-step guide with all the building blocks for creating an enterprise ready RAG application such as a question answering bot. We use a combination of different AWS services, open-source foundation models (FLAN-T5 XXL for text generation and GPT-j-6B for embeddings) and packages such as LangChain for interfacing with all the components and Streamlit for building the bot frontend.
We provide an AWS Cloud Formation template to stand up all the resources required for building this solution. We then demonstrate how to use LangChain for tying everything together:
- Interfacing with LLMs hosted on Amazon SageMaker.
- Chunking of knowledge base documents.
- Ingesting document embeddings into Amazon OpenSearch Service.
- Implementing the question answering task.
We can use the same architecture to swap the open-source models with the Amazon Titan models. After Amazon Bedrock launches, we will publish a follow-up post showing how to implement similar generative AI applications using Amazon Bedrock, so stay tuned.
Solution overview
We use the SageMaker docs as the knowledge corpus for this post. We convert the HTML pages on this site into smaller overlapping chunks (to retain some context continuity between chunks) of information and then convert these chunks into embeddings using the gpt-j-6b model and store the embeddings in OpenSearch Service. We implement the RAG functionality inside an AWS Lambda function with Amazon API Gateway to handle routing all requests to the Lambda. We implement a chatbot application in Streamlit which invokes the function via the API Gateway and the function does a similarity search in the OpenSearch Service index for the embeddings of user question. The matching documents (chunks) are added to the prompt as context by the Lambda function and then the function uses the flan-t5-xxl model deployed as a SageMaker endpoint to generate an answer to the user question. All the code for this post is available in the GitHub repo.
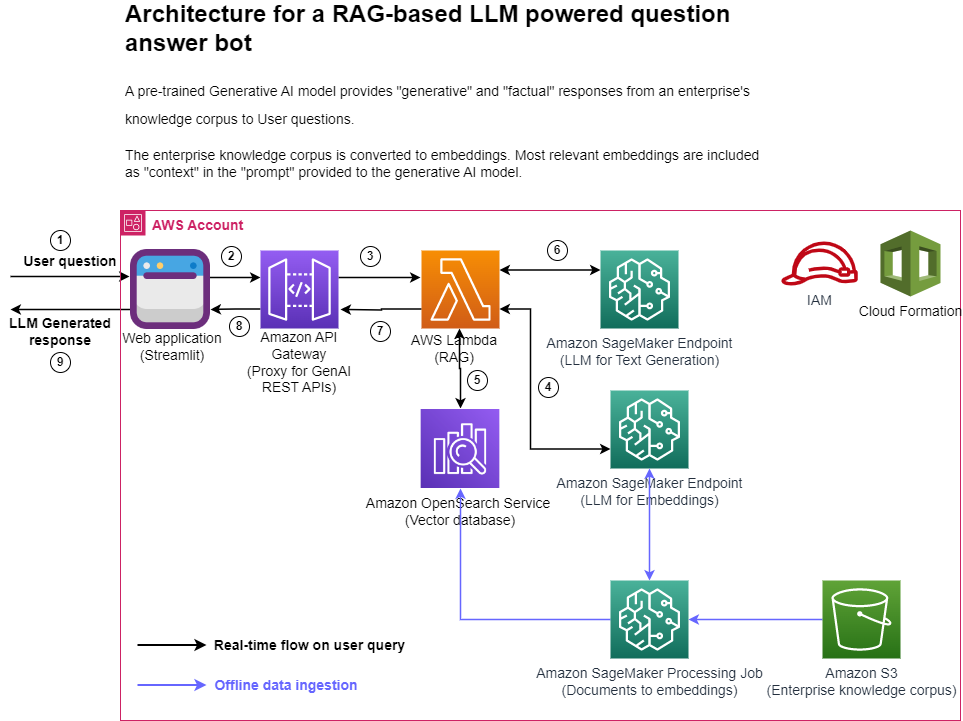
The following figure represents the high-level architecture of the proposed solution.
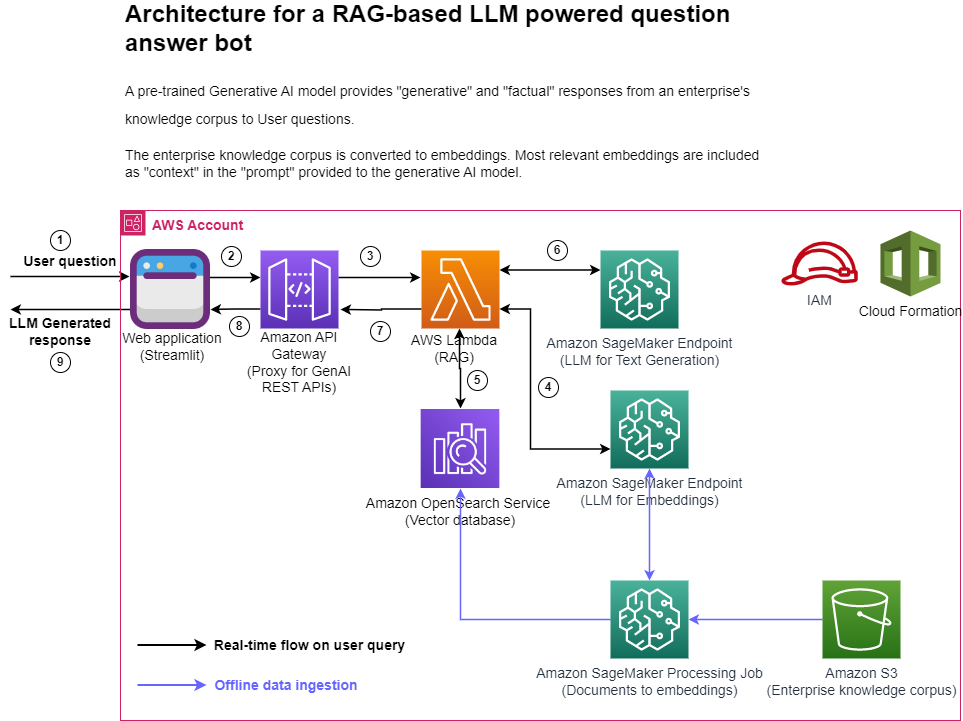
Figure 1: Architecture
Step-by-step explanation:
- The User provides a question via the Streamlit web application.
- The Streamlit application invokes the API Gateway endpoint REST API.
- The API Gateway invokes the Lambda function.
- The function invokes the SageMaker endpoint to convert user question into embeddings.
- The function invokes invokes an OpenSearch Service API to find similar documents to the user question.
- The function creates a “prompt” with the user query and the “similar documents” as context and asks the SageMaker endpoint to generate a response.
- The response is provided from the function to the API Gateway.
- The API Gateway provides the response to the Streamlit application.
- The User is able to view the response on the Streamlit application,
As illustrated in the architecture diagram, we use the following AWS services:
- SageMaker and Amazon SageMaker JumpStart for hosting the two LLMs.
- OpenSearch Service for storing the embeddings of the enterprise knowledge corpus and doing similarity search with user questions.
- Lambda for implementing the RAG functionality and exposing it as a REST endpoint via the API Gateway.
- Amazon SageMaker Processing jobs for large scale data ingestion into OpenSearch.
- Amazon SageMaker Studio for hosting the Streamlit application.
- AWS Identity and Access Management roles and policies for access management.
- AWS CloudFormation for creating the entire solution stack through infrastructure as code.
In terms of open-source packages used in this solution, we use LangChain for interfacing with OpenSearch Service and SageMaker, and FastAPI for implementing the REST API interface in the Lambda.
The workflow for instantiating the solution presented in this post in your own AWS account is as follows:
- Run the CloudFormation template provided with this post in your account. This will create all the necessary infrastructure resources needed for this solution:
- SageMaker endpoints for the LLMs
- OpenSearch Service cluster
- API Gateway
- Lambda function
- SageMaker Notebook
- IAM roles
- Run the data_ingestion_to_vectordb.ipynb notebook in the SageMaker notebook to ingest data from SageMaker docs into an OpenSearch Service index.
- Run the Streamlit application on a terminal in Studio and open the URL for the application in a new browser tab.
- Ask your questions about SageMaker via the chat interface provided by the Streamlit app and view the responses generated by the LLM.
These steps are discussed in detail in the following sections.
Prerequisites
To implement the solution provided in this post, you should have an AWS account and familiarity with LLMs, OpenSearch Service and SageMaker.
We need access to accelerated instances (GPUs) for hosting the LLMs. This solution uses one instance each of ml.g5.12xlarge and ml.g5.24xlarge; you can check the availability of these instances in your AWS account and request these instances as needed via a Sevice Quota increase request as shown in the following screenshot.
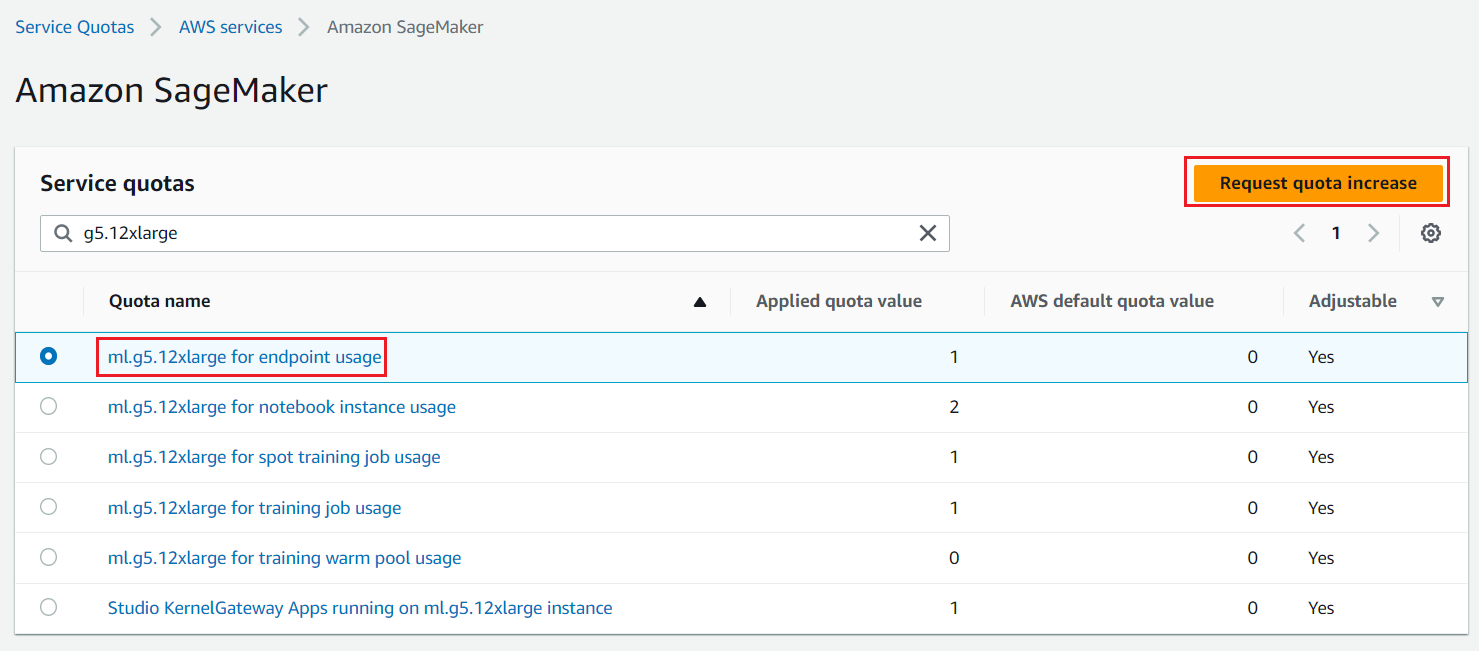
Figure 2: Service Quota Increase Request
Use AWS Cloud Formation to create the solution stack
We use AWS CloudFormation to create a SageMaker notebook called aws-llm-apps-blog and an IAM role called LLMAppsBlogIAMRole. Choose Launch Stack for the Region you want to deploy resources to. All parameters needed by the CloudFormation template have default values already filled in, except for the OpenSearch Service password which you’d have to provide. Make a note of the OpenSearch Service username and password, we use those in subsequent steps. This template takes about 15 minutes to complete.
| AWS Region | Link |
|---|---|
us-east-1 | |
us-west-2 | |
eu-west-1 | |
ap-northeast-1 |
After the stack is created successfully, navigate to the stack’s Outputs tab on the AWS CloudFormation console and note the values for OpenSearchDomainEndpoint and LLMAppAPIEndpoint. We use those in the subsequent steps.
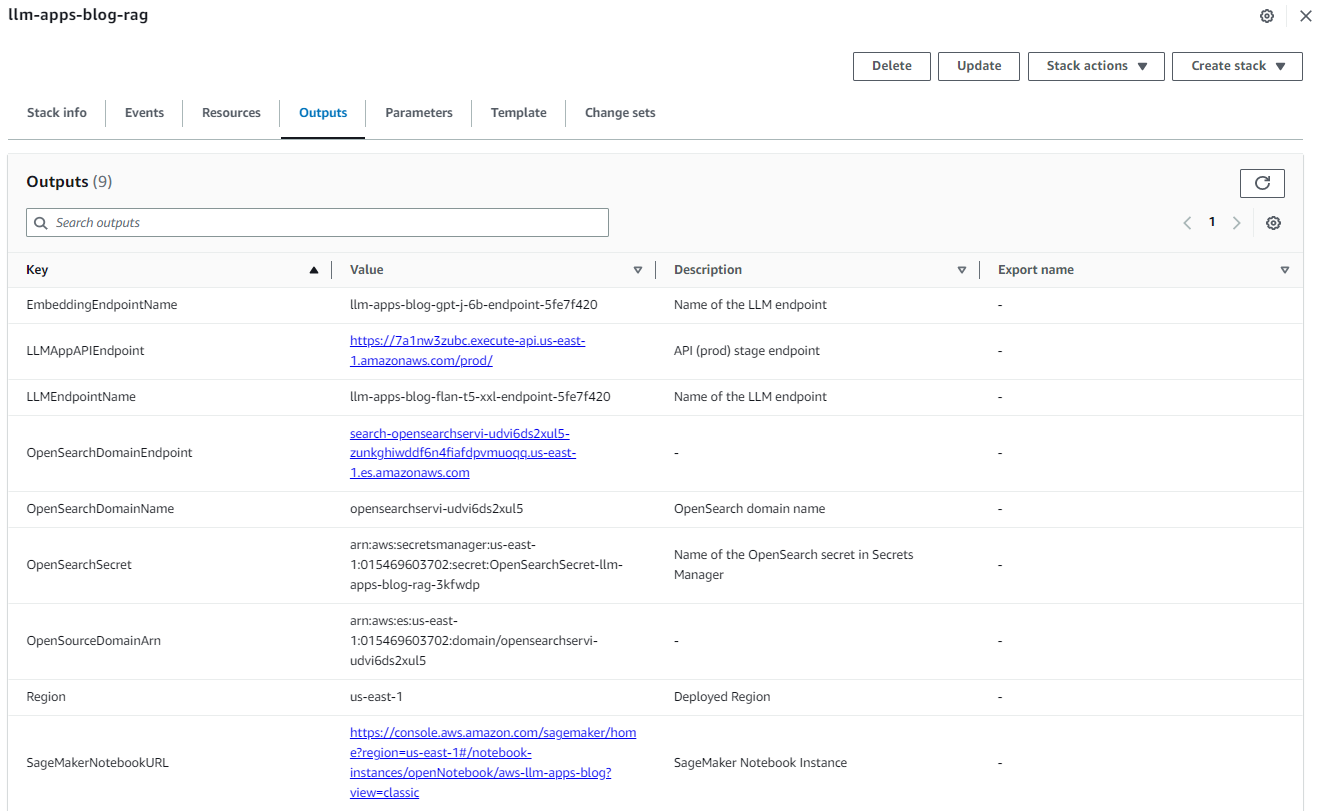
Figure 3: Cloud Formation Stack Outputs
Ingest the data into OpenSearch Service
To ingest the data, complete the following steps:
- On the SageMaker console, choose Notebooks in the navigation pane.
- Select the notebook
aws-llm-apps-blogand choose Open JupyterLab.
Figure 4: Open JupyterLab
- Choose data_ingestion_to_vectordb.ipynb to open it in JupyterLab. This notebook will ingest the SageMaker docs to an OpenSearch Service index called
llm_apps_workshop_embeddings.
Figure 5: Open Data Ingestion Notebook
- When the notebook is open, on the Run menu, choose Run All Cells to run the code in this notebook. This will download the dataset locally into the notebook and then ingest it into the OpenSearch Service index. This notebook takes about 20 minutes to run. The notebook also ingests the data into another vector database called FAISS. The FAISS index files are saved locally and the uploaded to Amazon Simple Storage Service (S3) so that they can optionally be used by the Lambda function as an illustration of using an alternate vector database.

Figure 6: Notebook Run All Cells
Now we’re ready to split the documents into chunks, which can then be converted into embeddings to be ingested into OpenSearch. We use the LangChain RecursiveCharacterTextSplitter class to chunk the documents and then use the LangChain SagemakerEndpointEmbeddingsJumpStart class to convert these chunks into embeddings using the gpt-j-6b LLM. We store the embeddings in OpenSearch Service via the LangChain OpenSearchVectorSearch class. We package this code into Python scripts that are provided to the SageMaker Processing Job via a custom container. See the data_ingestion_to_vectordb.ipynb notebook for the full code.
- Create a custom container, then install in it the LangChain and opensearch-py Python packages.
- Upload this container image to Amazon Elastic Container Registry (ECR).
- We use the SageMaker ScriptProcessor class to create a SageMaker Processing job that will run on multiple nodes.
- The data files available in Amazon S3 are automatically distributed across in the SageMaker Processing job instances by setting
s3_data_distribution_type='ShardedByS3Key'as part of theProcessingInputprovided to the processing job. - Each node processes a subset of the files and this brings down the overall time required to ingest the data into OpenSearch Service.
- Each node also uses Python multiprocessing to internally also parallelize the file processing. Therefore, there are two levels of parallelization happening, one at the cluster level where individual nodes are distributing the work (files) amongst themselves and another at the node level where the files in a node are also split between multiple processes running on the node.
- The data files available in Amazon S3 are automatically distributed across in the SageMaker Processing job instances by setting
- Close the notebook after all cells run without any error. Your data is now available in OpenSearch Service. Enter the following URL in your browser’s address bar to get a count of documents in the
llm_apps_workshop_embeddingsindex. Use the OpenSearch Service domain endpoint from the CloudFormation stack outputs in the URL below. You’d be prompted for the OpenSearch Service username and password, these are available from the CloudFormations stack.
The browser window should show an output similar to the following. This output shows that 5,667 documents were ingested into the llm_apps_workshop_embeddings index. {"count":5667,"_shards":{"total":5,"successful":5,"skipped":0,"failed":0}}
Run the Streamlit application in Studio
Now we’re ready to run the Streamlit web application for our question answering bot. This application allows the user to ask a question and then fetches the answer via the /llm/rag REST API endpoint provided by the Lambda function.
Studio provides a convenient platform to host the Streamlit web application. The following steps describes how to run the Streamlit app on Studio. Alternatively, you could also follow the same procedure to run the app on your laptop.
- Open Studio and then open a new terminal.
- Run the following commands on the terminal to clone the code repository for this post and install the Python packages needed by the application:
- The API Gateway endpoint URL that is available from the CloudFormation stack output needs to be set in the webapp.py file. This is done by running the following
sedcommand. Replace thereplace-with-LLMAppAPIEndpoint-value-from-cloudformation-stack-outputsin the shell commands with the value of theLLMAppAPIEndpointfield from the CloudFormation stack output and then run the following commands to start a Streamlit app on Studio. - When the application runs successfully, you’ll see an output similar to the following (the IP addresses you will see will be different from the ones shown in this example). Note the port number (typically 8501) from the output to use as part of the URL for app in the next step.
- You can access the app in a new browser tab using a URL that is similar to your Studio domain URL. For example, if your Studio URL is
https://d-randomidentifier.studio.us-east-1.sagemaker.aws/jupyter/default/lab?then the URL for your Streamlit app will behttps://d-randomidentifier.studio.us-east-1.sagemaker.aws/jupyter/default/proxy/8501/webapp(notice thatlabis replaced withproxy/8501/webapp). If the port number noted in the previous step is different from 8501 then use that instead of 8501 in the URL for the Streamlit app.
The following screenshot shows the app with a couple of user questions.

A closer look at the RAG implementation in the Lambda function
Now that we have the application working end to end, lets take a closer look at the Lambda function. The Lambda function uses FastAPI to implement the REST API for RAG and the Mangum package to wrap the API with a handler that we package and deploy in the function. We use the API Gateway to route all incoming requests to invoke the function and handle the routing internally within our application.
The following code snippet shows how we find documents in the OpenSearch index that are similar to the user question and then create a prompt by combining the question and the similar documents. This prompt is then provided to the LLM for generating an answer to the user question.
Clean up
To avoid incurring future charges, delete the resources. You can do this by deleting the CloudFormation stack as shown in the following screenshot.
Figure 7: Cleaning Up
Conclusion
In this post, we showed how to create an enterprise ready RAG solution using a combination of AWS service, open-source LLMs and open-source Python packages.
We encourage you to learn more by exploring JumpStart, Amazon Titan models, Amazon Bedrock, and OpenSearch Service and building a solution using the sample implementation provided in this post and a dataset relevant to your business. If you have questions or suggestions, leave a comment.
About the Authors
 Amit Arora is an AI and ML Specialist Architect at Amazon Web Services, helping enterprise customers use cloud-based machine learning services to rapidly scale their innovations. He is also an adjunct lecturer in the MS data science and analytics program at Georgetown University in Washington D.C.
Amit Arora is an AI and ML Specialist Architect at Amazon Web Services, helping enterprise customers use cloud-based machine learning services to rapidly scale their innovations. He is also an adjunct lecturer in the MS data science and analytics program at Georgetown University in Washington D.C.
 Dr. Xin Huang is a Senior Applied Scientist for Amazon SageMaker JumpStart and Amazon SageMaker built-in algorithms. He focuses on developing scalable machine learning algorithms. His research interests are in the area of natural language processing, explainable deep learning on tabular data, and robust analysis of non-parametric space-time clustering. He has published many papers in ACL, ICDM, KDD conferences, and Royal Statistical Society: Series A.
Dr. Xin Huang is a Senior Applied Scientist for Amazon SageMaker JumpStart and Amazon SageMaker built-in algorithms. He focuses on developing scalable machine learning algorithms. His research interests are in the area of natural language processing, explainable deep learning on tabular data, and robust analysis of non-parametric space-time clustering. He has published many papers in ACL, ICDM, KDD conferences, and Royal Statistical Society: Series A.
 Navneet Tuteja is a Data Specialist at Amazon Web Services. Before joining AWS, Navneet worked as a facilitator for organizations seeking to modernize their data architectures and implement comprehensive AI/ML solutions. She holds an engineering degree from Thapar University, as well as a master’s degree in statistics from Texas A&M University.
Navneet Tuteja is a Data Specialist at Amazon Web Services. Before joining AWS, Navneet worked as a facilitator for organizations seeking to modernize their data architectures and implement comprehensive AI/ML solutions. She holds an engineering degree from Thapar University, as well as a master’s degree in statistics from Texas A&M University.
Author: Amit Arora



