
Large-scale digital biomarker computation with AWS serverless services
This approach generates large amounts of data that requires processing… Digital biomarker data is typically saved in different formats, and various sources require different (and often multiple) processing steps… The key component in re-designing the pipeline is flexibility so the platform can…
Digital biomarkers are quantitative, objective measures of physiological and behavioral data. They are collected and measured using digital devices that better represent free-living activity in contrast to a highly structured in-clinic setting. This approach generates large amounts of data that requires processing.
Digital biomarker data is typically saved in different formats, and various sources require different (and often multiple) processing steps. In 2020, the Digital Sciences and Translational Imaging group at Pfizer developed a specialized pipeline using AWS services for analyzing incoming sensor data from wrist devices for sleep and scratch activity.
In time, the original pipeline needed updating to compute digital biomarkers at scale while maintaining reproducibility and data provenance. The key component in re-designing the pipeline is flexibility so the platform can:
- Handle various incoming data sources and different algorithms
- Handle distinct sets of algorithm parameters
These goals were accomplished with a framework for handling file analysis that uses a two-part approach:
- A Python package using a custom common architecture
- An AWS-based pipeline to handle processing mapping and computing distribution
This blog post introduces this custom Python package data processing pipeline using AWS services. The AWS architecture maintains data provenance while enabling fast, efficient, and scalable data processing. These large data sets would otherwise consume significant local resources.
Let’s explore each part of this framework in detail.
Custom Python package for data processing
Computation of digital biomarkers requires specific algorithms. Typically, these algorithms—for example, gait, activity, and more—had separate code repositories. Little thought was given to interaction and reproducibility.
Pfizer addressed this issue by creating the SciKit-Digital-Health Python package (SKDH) to implement these algorithms and data ingestion methods. As the SKDH framework is designed with reproducibility and integration in mind, modules can be chained together in a pipeline structure and run sequentially. Pipelines can be saved and loaded later; a key feature in enabling dynamic pipeline selection and reproducibility.
AWS framework for file analysis
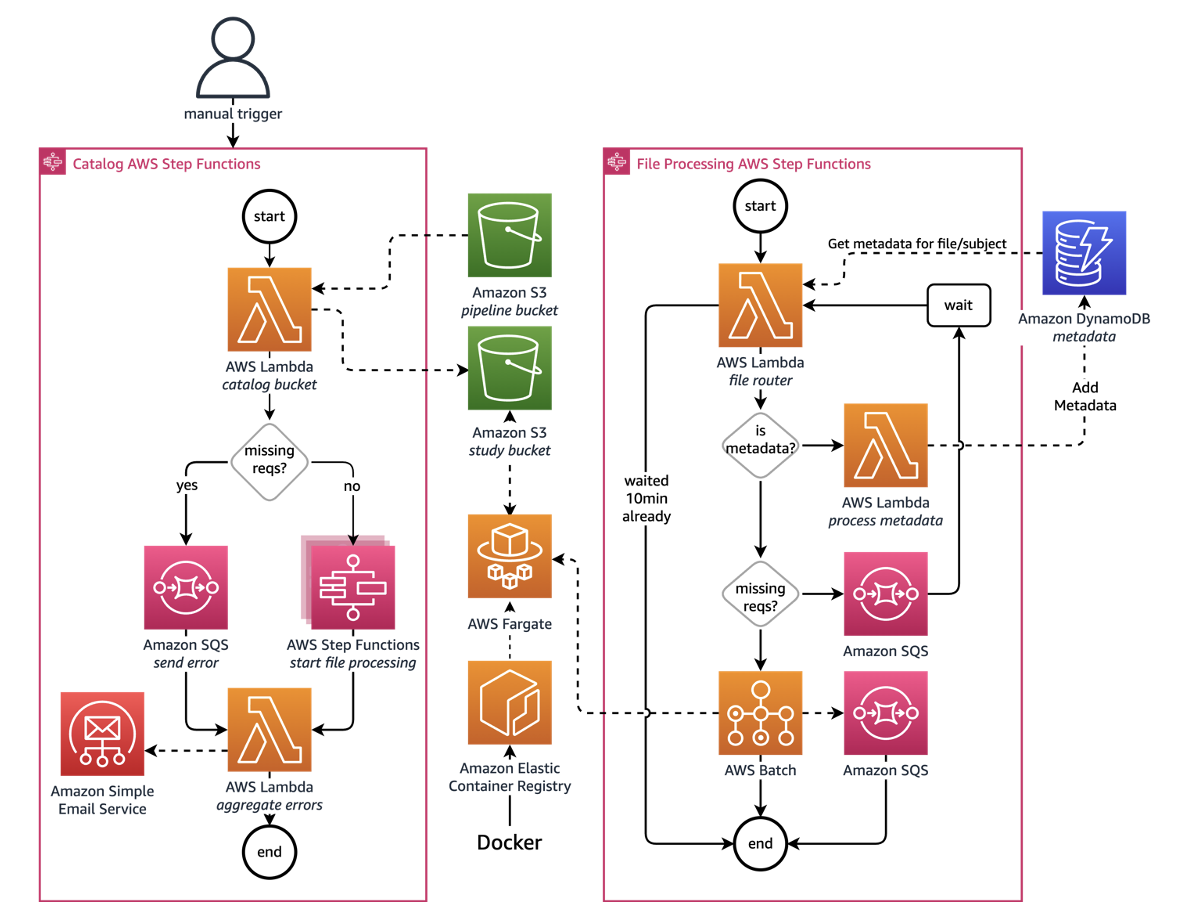
While SKDH is the core that computes the digital biomarkers, it requires an operational framework for handling all the data files. To that end, Pfizer and AWS designed the following architecture to handle file analysis on a study-by-study basis, as shown in Figure 1:
The overall platform can be broken into two components: the Catalog and File Processing AWS Step Functions workflows. Let’s explore each of these components, along with the necessary configuration file to process a study.
Catalog Step Functions workflow
The Catalog workflow searches study Amazon Simple Storage Service (Amazon S3) buckets for files that need processing. These files are defined in a study configuration document. The workflow consists of the following steps:
- The Catalog Step Function is manually triggered.
- If the study configuration file is found and the S3 Study buckets specified in the configuration file exist, search the S3 Study buckets for any files matching those enumerated in the study configuration file.
- If the configuration file or S3 Study bucket are not found, send a message to an Amazon Simple Queue Service (Amazon SQS) queue.
- For any cataloged files, start a new Processing workflow for each of these files.
File processing Step Functions workflow
The Processing workflow verifies processing requirements and processes both study data files and metadata files through the following steps:
- From the study configuration file, the workflow gets relevant details for the file. This includes metadata processing requirements as needed, and the specified set of SKDH algorithms to run.
- If the file contains metadata, a special AWS Lambda function cleans the data and saves it to the metadata Amazon DynamoDB table. Otherwise, check whether all requirements are met, including processing requirements (for example, height for gait algorithms) and logistics requirements (for example, file name information extraction functions).
- For missing requirements, the workflow will send a message to an Amazon SQS queue and try waiting for them to be met.
- If all requirements are met, it will start a batch job for the file.
- The batch job runs on AWS Fargate with a custom Docker image with a specified version of SKDH. This allows for easy versioning and reproducibility.
- The Docker image loads the specified SKDH pipeline and computes the digital biomarkers. Any results generated are then uploaded back to the S3 Study bucket at the end of processing.
Study configuration file
The configuration file defines everything necessary for processing a study. This includes which S3 bucket(s) the study data is located in, under which prefixes, and the list of file patterns of files to be processed. Figure 2 offers a sample configuration file example:
Building a stack with AWS CloudFormation
AWS CloudFormation provisions and configures all of the resources for the pipeline, allowing AWS services to be easily added and removed. CloudFormation integration with Github makes it easy to keep track of service changes. AWS Step Functions can also be integrated into CloudFormation template files, which gives an AWS specification in essentially one master file.
To iterate and update the stack, a combination of Python, Amazon SDK for Python (Boto3), and AWS Command Line Interface (AWS CLI) tools are used. Python allows for a more intricate command line script with positional and keyword arguments depending on the process. For example, updating the pipeline is a simple command, where <env> is the dev/stage/prod environment that needs to be updated:
Meanwhile, the Python function would be something like the following:
While it might seem unconventional to use both SDK for Python and a call to the AWS CLI from Python in the same script, the AWS CLI has some useful features that make the CloudFormation packaging and stack creation much easier. The end of the example file in Figure 3 pertains to the main utility in using the Python argparse package, which provides an easy to write user-friendly CLI.
This could also be achieved using AWS Cloud Development Kit (AWS CDK), an open source software development framework that allows you to define infrastucture in familiar programming languages. AWS CDK uses constructs that can be customized and reused.
Conclusion
This blog post shared a custom data processing pipeline using AWS services that, through intentional integration with a Python package, can run arbitrary algorithms to process data. By using AWS as the framework for this pipeline, data provenance is maintained while enabling fast, efficient, and scalable processing of large amounts of data that would typically consume significant company resources.
Author: Avni Patel




